Workflow management systems are as diverse as the business and scientific processes they support – from engineering to research to process automation. This article describes Nextflow – an open-source workflow manager widely used in life sciences. It covers the motivations behind Nextflow, explains what it is, and describes what the future holds for the platform.
Background and Motivations
Like similar open source efforts, Nextflow was born out of a need to solve specific problems while I was working as a research engineer in the Comparative Bioinformatics labs at the Centre for Genomics Regulation (CRG). Researchers were struggling with several issues that are all likely too familiar to workflows community members – complex, buggy scripts, long-running workflows that would suddenly fail, and challenges monitoring, managing, and maintaining workflows.
While there were several available workflow managers at the time, none of them specifically addressed our requirement. A fundamental challenge in the lab at that time was data handling. Comparative bioinformatics involves studying genome and protein sequences across species, and population-level studies can involve massive amounts of data. We needed a simple yet powerful framework to deploy the executions of thousands of tasks, comparing different alignment methods and protein sequences each other.
Some Key Ideas Behind Nextflow
Nextflow was designed from scratch having clear in mind key ideas and best practices:
- Allow developers to reuse any existing piece of software without the need of intermediate interface or wrapper; the tool command line is the interface and Linux is the integration layer.
- Manage tasks as a functional and self-contained unit of work. This was a key requirement to enable the deployment across heterogeneous computing platforms and allow auto-retry execution policy on failure.
- Provides a high-level abstraction for tasks parallelisation that allows developers to write simple yet high-scalable application, without the need to struggle with low level problem such as race conditions and locks to access shared resources.
- Strongly decouple the scientific application logic from the configuration & deployment setting, in order to streamline the deployment across different platforms and enable the migration to cloud environments.
- Remove unnecessary dependencies with external services and databases. We wanted a zero configuration experience both for the Nextflow runtime and the resulting pipelines.
- Enable debugging and recovering of failed executions. Bioinformatics pipeline can spin the execution of tens of thousands tasks. If something breaks we need a strategy that would allow the debugging of the failed task independently of the rest of the pipeline, and make it possible to recover the computation once the problem was solved from the last successfully computed tasks, to avoid throwing away days of computing resources.
Between these, very likely, the most important design choice that distinguishes Nextflow compared to other workflow management systems is the adoption of the data flow programming paradigm.
We often imagine workflows as a sequence of steps designed from the top down and including various dependencies, decision points, and sub-flows. There is another way to envision workflows, however, and that is from the perspective of the individual process steps. Individual steps have no notion of the overall flow. They have an input, perform some processing, and write data to an output, typically another step in the workflow.
Using this programming model you can think a workflow behaving like spreadsheet. In a spreadsheet, users enter expressions in cells that depend on calculations in other cells. The dependencies between cells can be complex, but users don’t think about how the spreadsheet will sequence calculations. Instead, they concentrate on the logic in each cell. When a cell changes, the spreadsheet worries about how to propagate changes to dependent cells. Nextflow is described as reactive because task execution is triggered when inputs change or become valid. It turns out that this model works surprisingly well at scale, and opportunities for parallelism occur naturally without the workflow designer needing to think about them.
From a technical point of view, Nextflow processes (aka tasks) can be thought of as reactive agents which run in parallel waiting for the input data that trigger their execution. It’s important to highlight that each of these processes are isolated from each other, and they can only communicate via asynchronous messages represented by dataflow variables.
Along with these, another pillar component of Nextflow was the adoption of containers as a core feature of the framework. Nextflow abstracts away the containerisation of the pipeline execution in a declarative manner. This means the user is only required to specify the container that needs to be used to run specific workflow tasks. Nextflow takes care to use this information to run the task within the container depending on the target execution platform that can be, for example, AWS Batch, an HPC batch scheduler e.g. Slurm or a local computer. This choice was proven to be critical to enable the portability and reproducibility of the resulting data analysis workflow. Nextflow nowadays supports multiple container runtime technologies, including Docker, Podman, Singularity, Charliecloud among others.
Nextflow Today
When Nextflow was launched as an open-source project in 2013, we couldn’t have imagined what it has become today. Today, Nextflow is downloaded over 55,000 times monthly and used by over 1,000 organizations, including some of the world’s largest pharmaceutical firms. Ideas like containers and support for source code managers (SCMs) are so thoroughly baked into Nextflow’s design that they seem commonplace.
In Nextflow, we were careful to decouple workflow logic from the details of underlying computing environments. To achieve this, Nextflow supports an abstraction called an Executor. Executors are pluggable components that enable pipelines to run without modification across virtually any compute environment, from a local host to an on-prem HPC cluster to various cloud services. Shifting to a new compute environment is as simple as changing a few lines of code in a configuration file.
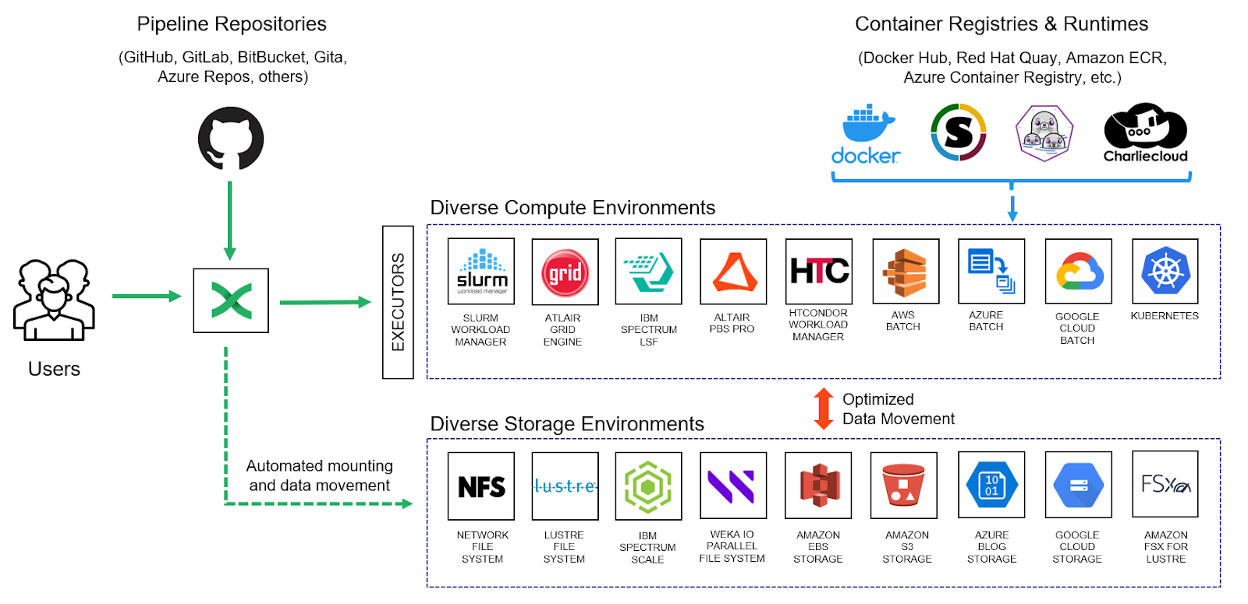
Pipelines can be stored locally or pulled from a preferred SCM at runtime. While applications can still be installed locally, pulling containers encapsulating bioinformatics tools has become the norm. Nextflow handles all the details, including compute resources, data movement, and making datasets visible to containers at runtime. Intermediate results and data are also cached, making flows resilient and recoverable after a failed step.
The nf-core Community
The nf-core community launched in 2018 marked a key milestone for the Nextflow community. nf-core is an independent effort to collect a curated set of analysis pipelines built using Nextflow. Significant effort has gone into developing tools, templates, and guidelines that enable domain experts to contribute to the community. The result is a set of high-quality pipelines that are portable, reproducible, fully documented, and cloud-ready. A recent State of the Workflow 2022 community survey showed that 62% of Nextflow users take advantage of these pipelines in their day-to-day research – a testament to their utility and the importance of this effort.
Towards the Future
We are fortunate to have vibrant user and developer communities actively engaged in Nextflow and helping to guide its evolution. We continue to support new computing environments, introduce new functionality, and improve throughput and scalability. I recently shared some plans for Nextflow in the article Evolution of the Nextflow runtime.
We are also planning to further enhance the support for containers providing a better automation on the overall container management lifecycle and ease the provisioning of multi-containers required by modern data analysis pipelines.
We are also improving our commercial Tower offering enabling improved collaboration, new data-related features, and features aimed at helping further optimize resource usage to reduce cloud spending.
You can learn more about Nextflow or download it for free by visiting nextflow.io. Existing Nextflow users can test-drive Nextflow Tower in the cloud at tower.nf.

 Paolo Di Tommaso (Nextflow creator and CTO of Seqera Labs)
Paolo Di Tommaso (Nextflow creator and CTO of Seqera Labs)